Hot Taq DNA Polymerase
 |

Description
Hot Taq DNA Polymerase is a thermostable recombinant DNA polymerase derived from thermophilic bacterium Thermus aquaticus. Its molecular weight is 94 kDa. Hot Taq DNA Polymerase can amplify DNA target up to 5 kb. The elongation velocity is 0.9~1.2kb/min. It has 5' to 3' polymerase activity but lacks of 3' to 5' exonuclease activity that results in a 3'-dA overhangs PCR product. All components of the PCR Buffer are at optimal concentration for efficient amplification. It contributes to highly specific incorporation of primer and template.
Applications
• PCR amplification of DNA fragments as long as 5 kb
• DNA labeling.
• DNA sequencing.
• PCR for cloning.
10X PCR Buffer with MgCl2
120mM Tris-HCl (pH 8.8), 500mM KCl, 1%Triton-X-100,
100mM Lycine, 25mM MgCl2
Storage Buffer
20mM TrisCl ( pH8.0), 100mM KCl, 3mM MgCl2 1mM DTTŁ¬0.1% NP-40 ,0.1% Tween20, 0.2mg/ml BSA, 50% (v/v) glycerol
Storage and Stability
The Hot Taq DNA Polymerase is shipped on Dry/Blue Ice. All kit components should be stored at -20ˇÉ upon receipt. Excessive freeze/thawing is not recommended. When stored under optimum conditions, the reagents are stable for a minimum of 6 months from date of purchase.
Quality Control
All preparations are assayed for contaminating endonuclease, exonuclease, and non-specific DNase activities. Functionally tested in DNA amplification.
Safety Precautions
Harmful if swallowed. Irritating to eyes, respiratory system and skin. Please refer to the material safety data sheet for further information.
Unit Definition
One unit is defined as the amount of the enzyme required to catalyze the incorporation of 10 nmole of dNTPs into an acid-insoluble form in 30 minutes at 70ˇĆC using hearing sperm DNA as substrate.
Note
• Hot Taq DNA Polymerase is for High Specificity PCR applications.
• The half-life of enzyme is > 40 minutes at 95ˇĆC.
• The error rate of Hot Taq DNA Polymerase in PCR is 2.2 x 10-5 errors per nt per cycler ; the accuracy (an inverse of the error rate) an average number of correct nucleotides in corporate before making an error, is 4.5 x 10-4 (determined according to the modified method described in).
• Hot Taq DNA Polymerase a accepts modified nucleotides (e.g. biotin-, digoxigenin-, fluorescent-labeled nucleotides) as substrates for the DNA synthesis.
• The number of PCR cycles depends on the amount of template DNA in the reaction mix and on the expected yield of the PCR product. 25–35 cycles are usually sufficient for the majority PCR reaction. Low amounts of starting template may require 40 cycles.
Figure
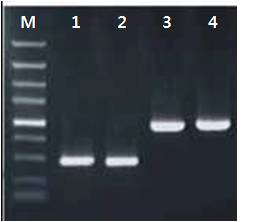
Amplification using Hot Start DNA Polymerase
M1: 100 bp DNA Ladder
1 and 2: 300bp amplification product generated using 0.2 mM dNTPs and 2.0u Hot start DNA Polymerase.
3 and 4: 5 kb and 8 kb amplification products generated using 0.25 mM dNTPs, 2.5u Hot start DNA Polymerase and 3% of formamide.
M2: Lambda / Hind III Marker
0.7% TAE agarose gel
M1: 100 bp DNA Ladder
1 and 2: 300bp amplification product generated using 0.2 mM dNTPs and 2.0u Hot start DNA Polymerase.
3 and 4: 5 kb and 8 kb amplification products generated using 0.25 mM dNTPs, 2.5u Hot start DNA Polymerase and 3% of formamide.
M2: Lambda / Hind III Marker
0.7% TAE agarose gel